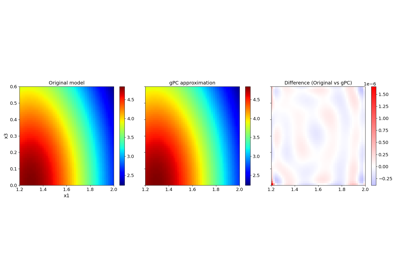
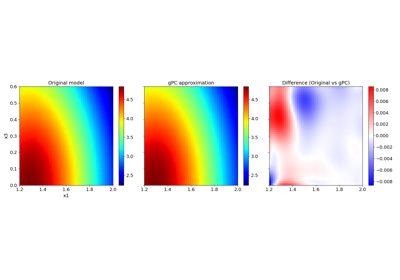
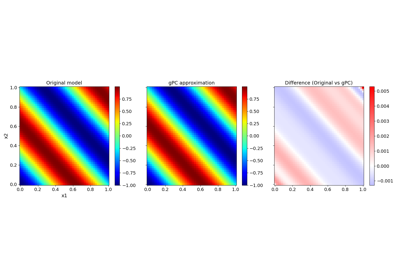
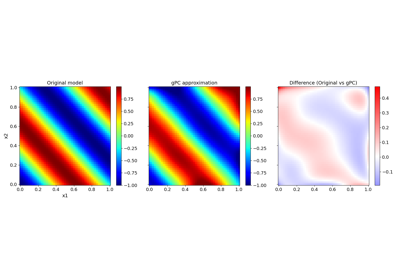
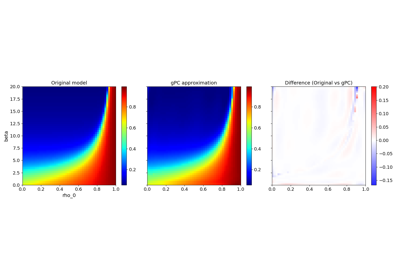
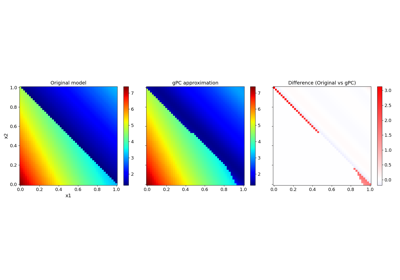
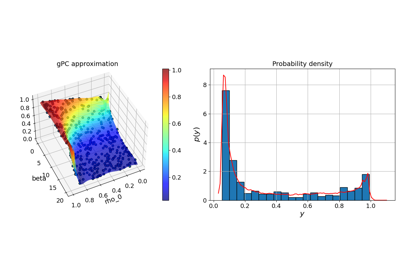
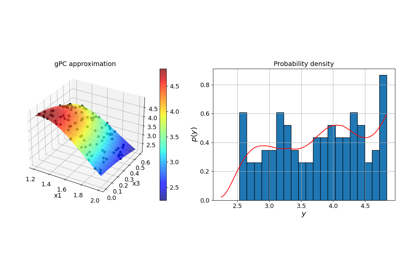
Basic features:#
Highly efficient uncertainty analysis of N-dimensional systems
Sensitivity analysis using Sobol indices and Global derivative based sensitivity indices
Easy coupling to user defined models written in Python, Matlab, Julia, etc…
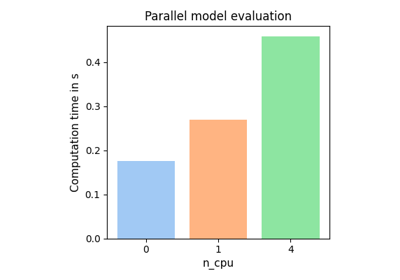
The parallelization concept allows to run model evaluations in parallel
Highly efficient adaptive algorithms allow for analysis of complex systems
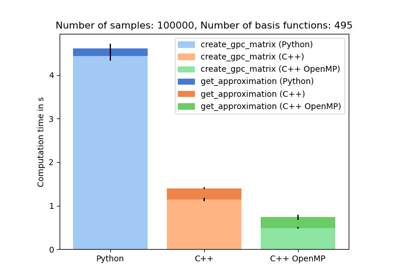
Includes highly efficient CPU and GPU (CUDA) implementations to significantly accelerate algorithmic and post-processing routines for high-dimensional and complex problems
- Includes state-of-the-art techniques such as:
Projection: determination of optimal reduced basis
L1-minimization: reduction of necessary model evaluations by making use of concepts from compressed sensing
Gradient enhanced gPC: use of gradient information of the model function to increase accuracy
Multi-element gPC: analyzing systems with discontinuities and sharp transitions
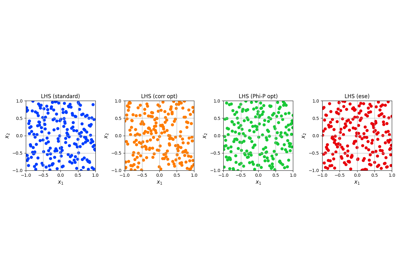
Optimized Latin Hypercube Sampling for fast convergence
Areas of application:#
pygpc can be used to analyze a variety of different of problems. It is used for example in the frameworks of:
Nondestructive testing:
Noninvasive brain stimulation:
Energy storage:
Aerospace engineering:
Cancer treatment:
If you use pygpc in your studies, please contact Konstantin Weise to extend the list above.
Installation#
Installation using pip:
Pygpc can be installed via the pip command with Python >= 3.6. Simply run the following command in your terminal:
pip install pygpc
If you want to use the plot functionalities of pygpc, please also install matplotlib and seaborn:
pip install matplotlib seaborn
Installation using the GitHub repository:
Alternatively, it is possible to clone this repository and run the setup manually. This requires a compiler that supports OpenMP which is used by the C-extensions and NumPy for some headers. You can install NumPy by running the following command:
pip install numpy
Alternatively you can install the build dependencies with the following command:
pip install -r requirements.txt
Afterwards, pygpc can be installed by running the following line from the directory in which the repository was cloned:
python setup.py install
Installation of the CUDA backend:
Pygpc also provides a CUDA-backend to speed up some computations. To use the backend you need to build it manually. This requires the CUDA-toolkit and CMake. CMake can be installd via the pip command. Simply run the following command in your terminal:
pip install cmake
For the installation of the CUDA-toolkit please refer to: https://docs.nvidia.com/cuda/cuda-quick-start-guide/index.html. If CMake and the CUDA-toolkit are installed on your machine you can build the extension with:
python build_pygpc_extensions_cuda.py
Troubleshooting for OSX:
On a mac you need GCC to install pygpc. If you are using the brew package manager you can simply run:
brew install gcc libomp
Then install pygpc with:
CC=gcc-9 CXX=g++-9 python setup.py install
Troubleshooting for Windows:
On windows you might need a compiler to install pygpc. To install the Visual C++ Build Tools, please refer to: http://go.microsoft.com/fwlink/?LinkId=691126&fixForIE=.exe.
Documentation#
For a full API of pygpc, see https://pygpc.readthedocs.io/en/latest/.
For examplary simulations and model configurations, please have a look at the jupyter notebooks provided in the /tutorial folder and the templates in the /templates folder.
Reference#
If you use pygpc, please cite:
Contact#
If you have questions, problems or suggestions regarding pygpc, please contact Konstantin Weise.
Examples Gallery#
- orphan:
Introduction#

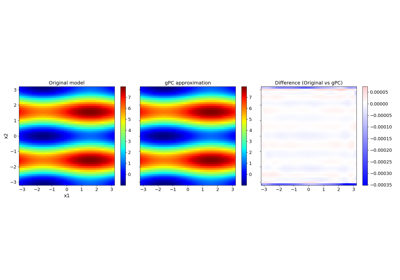
Introduction to generalized Polynomial Chaos (gPC)
Gallery generated by Sphinx-Gallery
- orphan:
Generalized Polynomial Chaos#
Gallery generated by Sphinx-Gallery
- orphan:
Algorithms#
Gallery generated by Sphinx-Gallery
- orphan:
Sampling schemes#
Gallery generated by Sphinx-Gallery
- orphan:
Features#
Gallery generated by Sphinx-Gallery
- orphan:
Examples#
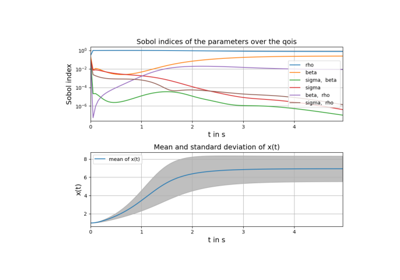
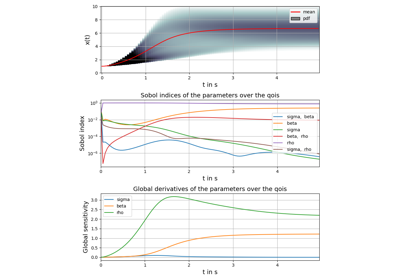
Example: Lorenz system of differential equations (Julia)